Expertise and instruments
AG Fakler
AG Fakler
Institute of Physiology II
Albert-Ludwigs-University
Hermann-Herderstr. 7
79104 Freiburg
Email: bernd.fakler@physiologie.uni-freiburg.de
Tel: +49 761 203 5175
All instruments are equipped with nano-HPLC-Systems and Nano-spray ionization units.
The central goal of our research is to understand the molecular principles underlying fast signaling at and across the plasma membrane. Speed and functional specificity of this signaling are primarily encoded by protein-protein interactions that give rise to protein (super)complexes, networks and (sub)organellar compartments.
To characterize these higher-order assemblies in native cells and tissues we have established three functional proteomic platforms
Our MS-based analyses use either label-free quantification of protein amounts on in-house developed procedures and algorithms, or quantification based on stable isotope labelling (e.g. SILAC). In addition, we pursue exploratory projects on the identification of post-translational modifications of and small molecule ligand binding to selected target proteins. Our in-house MS facility operates with three hybrid tandem mass spectrometers (Orbitrap XL, Orbitrap Elite, QExactive HF-X, each coupled to a nano-HPLC system) covering the specific requirements of our projects with respect to cycling/sequencing speed, sensitivity and mass accuracy.
Bildl W, et al. Extending the dynamic range of label-free mass spectrometric quantification of affinity purifications. Mol Cell Proteomics 11, M111 007955 (2012)
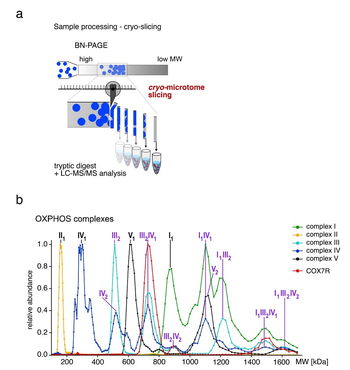
Muller CS, et al. Cryo-slicing Blue Native-Mass Spectrometry (csBN-MS), a Novel Technology for High Resolution Complexome Profiling. Mol Cell Proteomics 15, 669-681 (2016).
Muller CS, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci 107, 14950-14957 (2010).
Schwenk J, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74, 621-633 (2012).
Schwenk J, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313-1319 (2009).
Schwenk J, et al. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature 465, 231-235 (2010).
Schwenk J, et al. Modular composition and dynamics of native GABAB receptors identified by high-resolution proteomics. Nat Neurosci 19, 233-242 (2016).